Microbial Source Tracking in Vulnerable Water Resources
29 Dic 2019
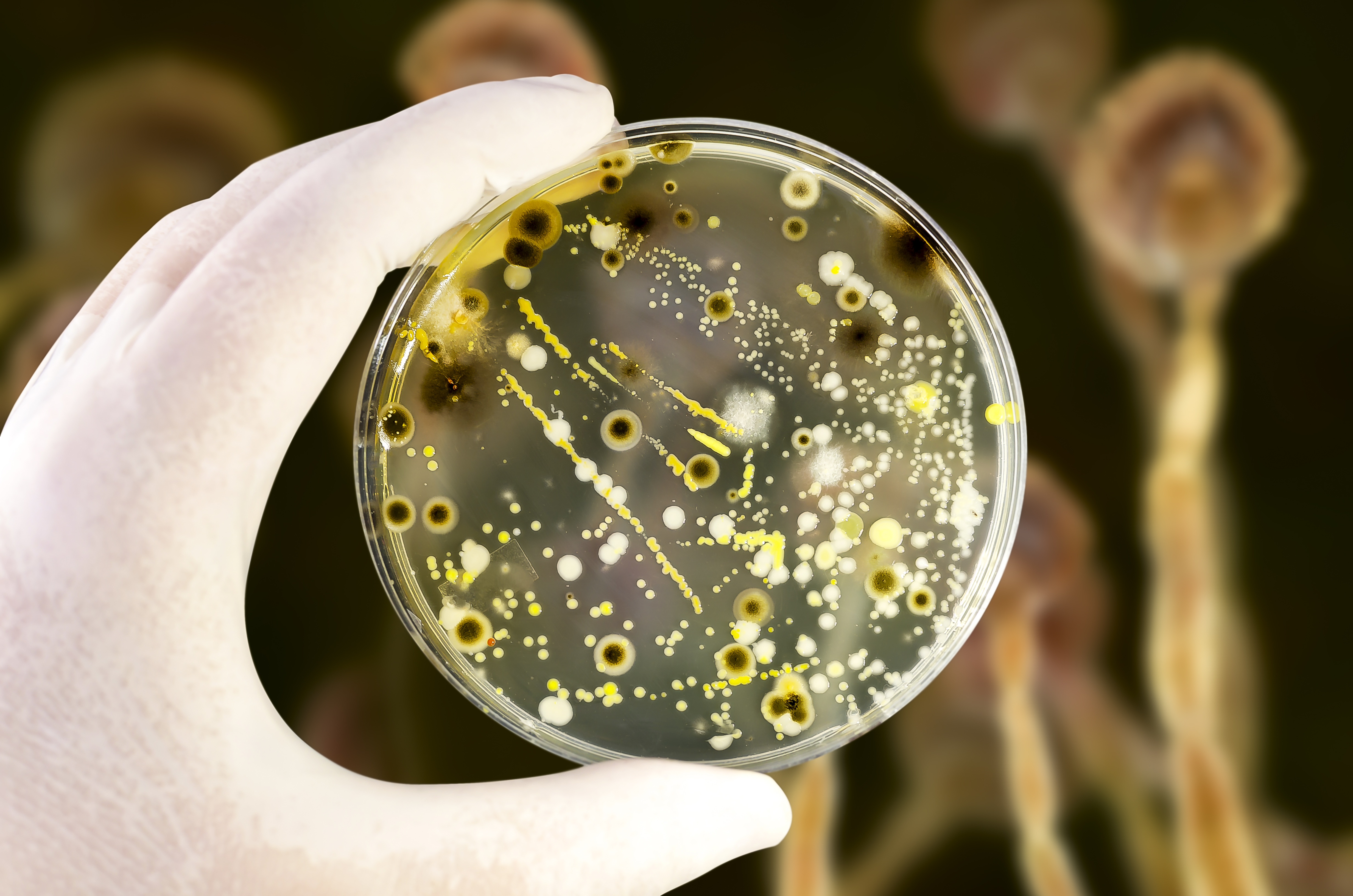
Areas with karst geology are often major sources of drinking water, however, that water is at risk from contamination.
Fractures, sinkholes and other discontinuities enable the rapid movement of surface water into underground aquifers, which have short water retention times. Rainfall-driven transfer of animal and human faecal pollution from the surface into aquifers is a known problem with incidents reported.1 Microbial source tracking (MST) techniques can be used to discriminate among different sources of faecal contamination of water, leading to targeted environmental management interventions. Culture-dependent Bacteroides phage and molecular-dependent Bacteroidales 16S rRNA assays are employed in MST studies globally.
One of the most promising set of tools to emerge from the MST toolbox has been a suite of assays based on the polymerase chain reaction (PCR). PCR assays detect and amplify specific DNA sequences, producing banding patterns and other signals which can be detected and quantified by various means. A range of microorganisms is present in faeces, which are specific to their animal hosts. Total DNA is extracted from a water sample and the sample is examined using PCR for DNA from source-specific organisms. The presence of certain microorganisms indicates the source of the faecal contamination. Assays specific for humans, herbivores, dogs and wildfowl are in development.
The Bacteroides-Prevotella group of bacteria resides exclusively in the gut of warm-blooded animals. They are strict anaerobes, which means they are highly unlikely to replicate in the environment. They are excreted in higher numbers than the faecal coliforms and therefore a universal PCR marker designed to detect all of the members of the Bacteroides-Prevotella group (termed Total Bacteroidetes) in waterways is a useful indicator of the presence of faecal pollution.
In an alternative approach, individual colonies, usually of E. coli or enterococci, are isolated from water samples. These isolates are then fingerprinted using either DNA-based or phenotypic/biochemical based methods to generate an indivdiual profile of each isolate, which is compared with the profiles of isolates in a library of known faecal sources. The library is usually comprised of E. coli or enterococci isolates collected from potential sources of contamination such as human sewage or animal faeces, located in the same environment as the water sample.
Each one of these tools is relevant for a particular purpose and when used in conjunction with other tools, can increase the certainty that pollution is derived from a specific source. Diston et al.2 used both types of approach for a detailed study of two water sources in a karst area of Switzerland. For blank and positive standards it is essential to have a high purity water supply. They used sterile ultrapure water from an ELGA PURELAB Ultra. They confirmed that sites representing karst aquifers were of significantly worse microbial quality than those from other hydro-geological settings.
References
- P. Beaudeau, H. de Valk, V. Vaillant, C. Mannschott, C. Tillier, D. Mouly and M. Ledrans, “Lessons learned from ten investigations of waterborne gastroenteritis outbreaks, France, 1998-2006”, J. Water Health, 6,(4) 491-503, 2008
- D. Diston, R. Robbi, A. Baumgartner and R. Felleisen, „Microbial source tracking in highly vulnerable karst drinking water resources“, Journal of Water and Health, 16.1, 138-149, 2018
Dr Paul Whitehead
After a BA in Chemistry at Oxford University, Paul focused his career on industrial applications of chemistry. He was awarded a PhD at Imperial College, London for developing a microwave-induced-plasma detector for gas chromatography. He spent the first half of his career managing the analytical support team at the Johnson Matthey Research/Technology Centre,specialising in the determination of precious metals and characterising applications such as car-exhaust catalysts and fuel cells. Subsequently, as Laboratory Manager in R&D for ELGA LabWater, he has been involved in introducing and developing the latest water purification technologies. He now acts as a consultant for ELGA.
